SeraMir Exosome RNA Column Purification Kit
Overview
Efficient RNA extraction from exosomes with SBI’s most flexible SeraMir kit
When you want to extract RNA from already isolated exosomes, choose the SeraMir Exosome RNA Column Purification Kit. With only SBI’s highly efficient SeraMir RNA purification columns and reagents, you can keep your exosome isolation and downstream RNA processing options open.
How It Works
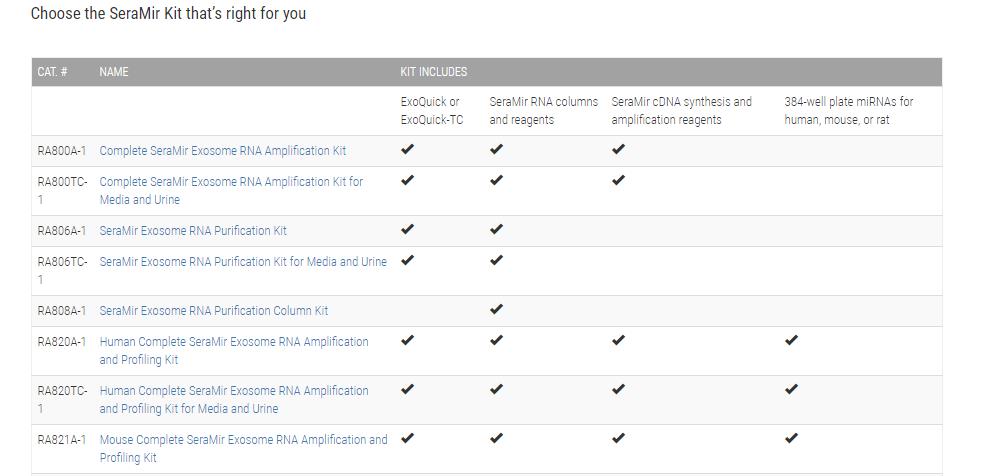
Choose the SeraMir Kit that’s right for you
Simply add the phenol-free exosome lysis buffer to your isolated exosomes and then run through the SeraMir RNA purification columns. In just three minutes, your highly pure exoRNAs will be ready for the next step, whether it’s processing for qPCR, microanalysis, NGS, or another application.

Supporting Data
Better qPCR profiling with SeraMir

Figure 1. Serum RNA prepared by the SeraMir Kit delivers more reliable, reproducible qPCR profiles than when the RNA is isolated using conventional Trizol methods. Profiling of 380 Human microRNAs across the SeraMir 384 Profiler. The phenol-free exosome lysis step coupled to the small RNA binding columns isolates exoRNAs with much higher purity than Trizol/Phenol based methods. The SeraMir exoRNAs are compatible with downstream polyadenylation and reverse trancription reactions for amplification and accurate qPCR profiling.

Figure 2. Serum exoRNAs prepared using SeraMir deliver excellent performance in microarray studies. Samples from a pooled normal serum preparation and from a male caucasian (age 73) with adenocarcinoma of the colon were used in this study. Exosomes were precipitated from 250 µL of serum using the SeraMir Exosome RNA Amplification Kit. The T7-amplified “sense” exoRNAs were then used for direct labeling analyses on LC Sciences miRBase ver.16 array chips (performed in triplicate). The exoRNAs were hybridized across 1,214 different microRNAs on the probe set.
Of the 1,214 microRNAs analyzed, 79 microRNAs showed a signal intensity >32. Within this set of 79, there was a clear colon versus normal “signature set” of 40 microRNAs that could discriminate normal from colon cancer serum samples with a p-value < 0.01. The identities of the microRNAs found in this study have been masked while further investigation continues.
Citations
-
Zhou, X, et al. (2017) Characterization of mouse serum exosomal small RNA content: The origins and their roles in modulating inflammatory response. Oncotarget. 2017 Jun 27; 8(26):42712-42727. PM ID:28514744
-
Zhang, R, et al. (2017) Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun.. 2017 Jun 13;. PM ID: 28623135
-
Huleihel, L, et al. (2017) Matrix bound nanovesicles recapitulate extracellular matrix effects on macrophage phenotype. Tissue Eng Part A. 2017 Jun 4;. PM ID: 28580875
-
Holliday, LS, et al. (2017) Exosomes: novel regulators of bone remodelling and potential therapeutic agents for orthodontics. Orthod Craniofac Res. 2017 Jun 1;:95-99. PM ID: 28643924
-
Koyama, S, et al. (2017) Dynamic changes of serum microRNA-122-5p through therapeutic courses indicates amelioration of acute liver injury accompanied by acute cardiac decompensation. ESC Heart Fail. 2017 May 1; 4(2):112-121. PM ID: 28451447
-
Selmaj, I, et al. (2017) Global Exosome Transcriptome Profiling Reveals Biomarkers for Multiple Sclerosis. Ann. Neurol.. 2017 Apr 15;. PM ID: 28411393
-
Val, S, et al. (2017) Purification and characterization of microRNAs within middle ear fluid exosomes: implication in otitis media pathophysiology. Pediatr. Res.. 2017 Apr 5;. PM ID: 28157838
-
Faust, A, et al. (2017) Urinary bladder extracellular matrix hydrogels and matrix-bound vesicles differentially regulate central nervous system neuron viability and axon growth and branching. J Biomater Appl. 2017 Apr 1; 31(9):1277-1295. PM ID: 28447547
-
Tsukita, S, et al. (2017) MicroRNAs 106b and 222 Improve Hyperglycemia in a Mouse Model of Insulin-Deficient Diabetes via Pancreatic β-Cell Proliferation. EBioMedicine. 2017 Feb 1; 15:163-172. PM ID:27974246
-
Hubal, MJ, et al. (2017) Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obesity (Silver Spring). 2017 Jan 1; 25(1):102-110. PM ID:27883272
-
Nakano, T, et al. (2017) Hepatic miR-301a as a Liver Transplant Rejection Biomarker? And Its Role for Interleukin-6 Production in Hepatocytes. OMICS. 2017 Jan 1; 21(1):55-66. PM ID: 28271982
-
Peng, Y, Huleihel, L & Cardenes, N. (2017) Different MiroRNA Expression In MSC-Derived Exosomes: IPF Patients And Age-Matched Normal Individuals. Conference. ;. Link: Conference
-
Protocol, IIEI. (2017) List of Components. Product. ;. Link: Product
-
Josson, S, Gururajan, M & WKChung, L. (2017) miRNA Characterization from the Extracellular Vesicles. bio-protocol. ; 7(4). Link: bio-protocol
-
Sueta, A, et al. (2017) Abstract P1-02-13: Differential expression of exosomal miRNAs between breast cancer patients with recurrence and no-recurrence. Abstract. ;. Link: Abstract
-
Reza, AM, et al. (2016) Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci Rep. 2016 Dec 8; 6:38498. PM ID: 27929108
-
Liu, C, et al. (2016) Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget. 2016 Nov 15; 7(46):76250-76260. PM ID: 27788488
-
Nakamura, K, et al. (2016) Altered expression of CD63 and exosomes in scleroderma dermal fibroblasts. J. Dermatol. Sci.. 2016 Oct 1; 84(1):30-39. PM ID: 27443953
-
Gutierrez, MJ, et al. (2016) Airway Secretory microRNAome Changes during Rhinovirus Infection in Early Childhood. PLoS ONE. 2016 Sep 20; 11(9):e0162244. PM ID: 27643599
-
Huang, Y, et al. (2016) 血清中外泌体及外泌体 RNA 提取方法的比较. 中华检验医学杂志. 2016 Jul 20; 39:427-432. Link: 中华检验医学杂志
